RCloud is open-source software developed primarily by Simon Urbanek and Gordon Woodhull of AT&T Research and Carlos Scheidegger, University of Arizona to facilitate collaboration for big data analytics and visualizations. You can test drive the most recent release of RCloud by logging in as a Data Scientist using our Try It page, or you can create a local private RCloud; detailed instructions are provided in our Public versus Private (Download / Local Installation) RClouds tutorial.
Traditional Data Science environments often assume a single-developer perspective, while teams of data scientists often benefit from leveraging existing analyses, built-in version control, automated recommendations, and easier sharing of code, data, annotations and comments.
RCloud provides a notebook interface that lets you easily generate a web-based session, annotate it with text, equations, supporting images and web page layout so that data scientist teams can convert collaborative work on large complex datasets into reports and web pages (dashboards) which may easily be shared with others.
The following paragraphs provide a bried overview of some of the unique features of RCloud, but more detailed feature explanations are provided in the “WHAT’S NEW” and “GUI DOCUMENTATION” sub-menu pages for each release and more detailed technical explanations are provided in the RCloud Wiki and in our RCloud API.
The RCloud ecosystem is a organized around notebooks. A notebook is a collection of R or Python scripts together with data and code assets which the scripts may rely on. Assets may be data files, like CSV or Hierarchical Data Format (HDF) files, image files, which are displayed as part of a user-interface, or HTML, CSS and Javascript code.
All of the elements of the notebook are stored on a Github server. The Github server supports continual and automatic background versioning of notebooks as they are developed and modified. This feature provides the benefits of basic Git version control for casual users and the option to use more sophisticated git features for advanced users.
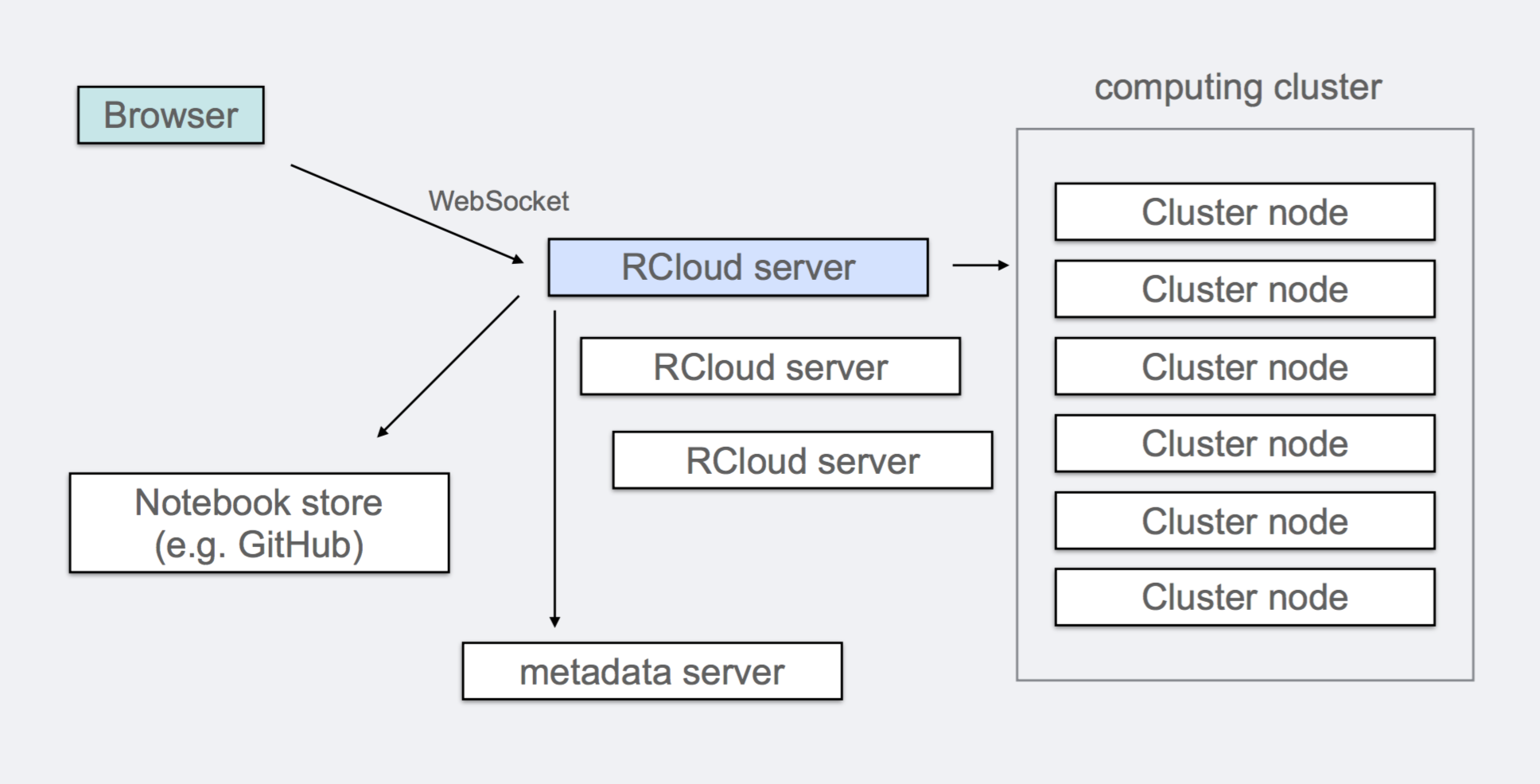
The RCloud editor is designed as an integrated development environment (IDE) for creating scripts to be executed and shared by internal Data Scientists and potentially deployed for execution by any web user. The basic architecture of the system is shown in the figure below.
The the system back-end depicts an RCloud server which executes code entered at the web-browser and communicates the result back to the browser.
RCloud was specifically designed to leverage existing systems and standards so communication between most parts of the system happens through HTTP. As a result, some desirable features of RCloud become natural, such as native support for RCloud notebooks as web services.
Every notebook in RCloud is named by a URL and notebooks by default are visible in the entire organization. This is deliberate as broad access to analysis outputs increases long-term engagement in part through cross-references on the web.
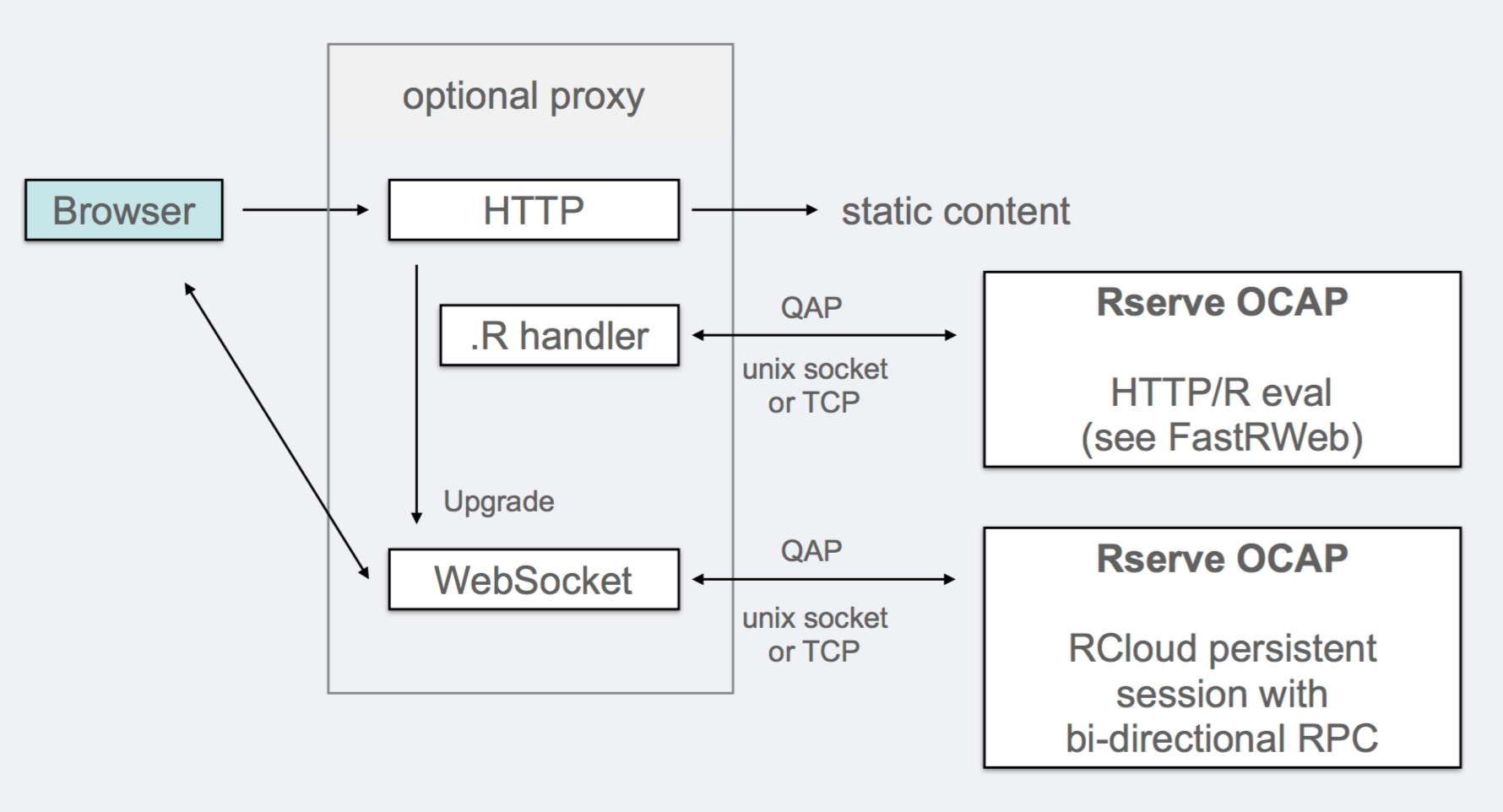
The security challenge in a system like this is to allow a given internal Data Scientist access to the full power of the R interpreter on the server while preventing external users from executing their own arbitrary code. RCloud has adopted the discipline known as Object-Capabilities as outlined by Mark Miller, Robust Composition: Towards a Unified Approach to Access Control and Concurrency Control (2006) [PDF] to arbitrate the communication of functional information between the web-browser and the R interpreter.
Object-capabilities support the notion of mapping language functions to cryptographic hashes which act as function proxies on the exposed client. The server then acts on these hashes rather than on direct code, thereby preventing the client from submitting arbitrary code to the server.
RCloud uses the RServe protocol to implement the Object-Capabilities methodology and the client-server communication. An RCloud session runs on both client and server and it is possible for R functions on the server to call JavaScript functions on the client and vice versa; see [RCloud application program interface (API) and R and Javascript binding] (https://github.com/att/rcloud/wiki/R-JavaScript-binding) for a detailed description of the browser/server interaction.
Individual notebooks are protected with privacy controls. Notebooks may be protected through group assignments, made entirely private (readable only by you), or made public (readable by anyone). Unlike a hidden notebook, protected notebooks are not readable by anyone without permission, even within your GitHub instance.
RCloud notebooks may also be deployed as a web-service provider via the notebook.R interface. This interface allows execution of any RCloud notebook, or access to any notebook asset, by simply referencing the associated URL. This feature also allows RCloud notebooks to be integrated with other web-technologies simply be referencing them.
For example, to generate a dynamic web browser image, create a web page with the following HTML <img> element.
<img width=300, height=300, src="https://rcloud.social/notebook.R/rclouddocs/RCloud%20Sample%20Notebooks/Dashboarding/RCloud%20notebook.R/foo.png" >Then as user ‘rclouddocs’ create a notebook named ‘foo.png’ containing the following plotting code:
run = function(...)
{
p = WebPlot(300,300)
plot(runif(10),runif(10))
p
}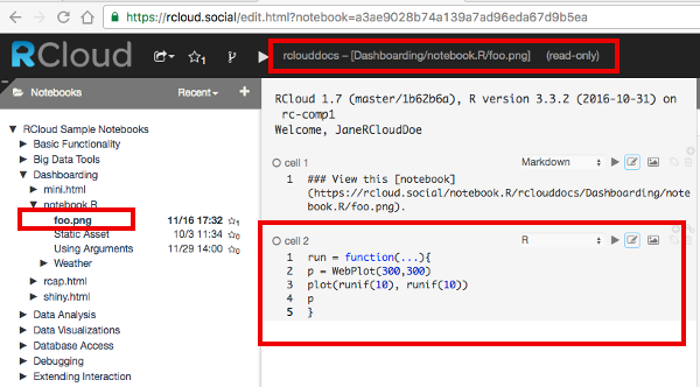
In this example the RCloud server will intercept the HTTP request for the the <img> source file and return the result of the notebook, a PNG graphics file, which will be displayed at the location of the <img> element.
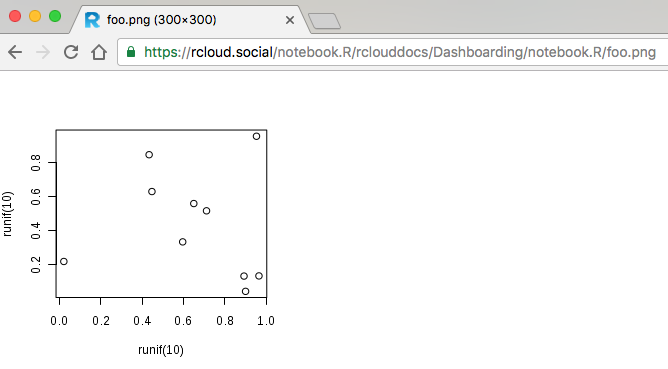
The WebPlot() function used in this example is from the FastRWeb [PDF] R package which includes several other useful functions for creating web content with R. Our Weather Dashboard illustrates how the output from individual notebook.R_ RCloud notebooks may be used to create dashboards.
Notebooks can be executed one cell at a time in an interactive session, similar to traditional read-eval-print loops, or can all be executed concurrently, similar to running a shell script. One of the main contributions of RCloud is the notion that notebooks are “always deployed”.
In other words, the most recent version of a notebook is immediately available to all other users of the system. Another way to describe this is that RCloud lacks a “save” button: any notebook cell that runs is always associated to a notebook version serialized to disk.
For more about the goals, architecture and design of RCloud see Stephen North, et. al., Collaborative Visual Analysis with RCloud (2013) [PDF]